Higher throughput volume EM via integrated array tomography
- Abstract number
- 1081
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.1081
- Corresponding Email
- [email protected]
- Session
- LST.4 - Volume Scanning Electron Microscopy in life sciences
- Authors
- Ryan Lane (1), Anouk Wolters (2), Ben Giepmans (2), Jacob Hoogenboom (1)
- Affiliations
-
1. TU Delft
2. UMC Groningen
- Keywords
Array tomography, correlative light electron microscopy, fluorescence microscopy, integrated microscopy, throughput, volume electron microscopy
- Abstract text
Fuelled by the rapidly expanding field of connectomics, volume electron microscopy (EM) has experienced an explosive growth in the past decade [1]. Due to the inherently slow pace of electron-probe-based scanning techniques, however, it can take years to acquire the vast datasets necessary to trace neurons throughout the brain. Recent approaches to enhancing throughput of volume EM have primarily fallen into one of two camps. Multi-beam approaches image an entire volume by using multiple beams in parallel (either within a single instrument [2], [3] or across multiple instruments [4]). Multi-scale approaches reduce the total imaging volume by first identifying regions of interest (ROI) from either the same [5] or a different [6], [7] imaging modality (so-called multi-modal approaches) at lower magnification. Though they are capable of realizing tremendous gains in throughput, the cost and technical challenges associated with multi-beam instrumentation can be prohibitively expensive. Multi-modal approaches, on the other hand, often present their own challenges with respect to ROI tracking and correlation across modalities. Here we present a multi-modal technique for volume EM that circumvents these challenges by integrating a fluorescence microscope into the chamber of a conventional SEM.
Fluorescence microscopy (FM) is an imaging technique widely sought out in cell biology for its use in finding rare events or localizing particular hormones or organelles of interest. When combined with the ultrastructural information provided by EM, functional information can be correlated with composition, offering profound insight to many biological questions. Hence, by using FM as a guide within the integrated microscope, it is possible to seamlessly identify, navigate to, and image regions of interest with high resolution EM. We demonstrate this process of integrated array tomography on 80nm rat pancreas tissue and show its viability for high precision, volume CLEM.
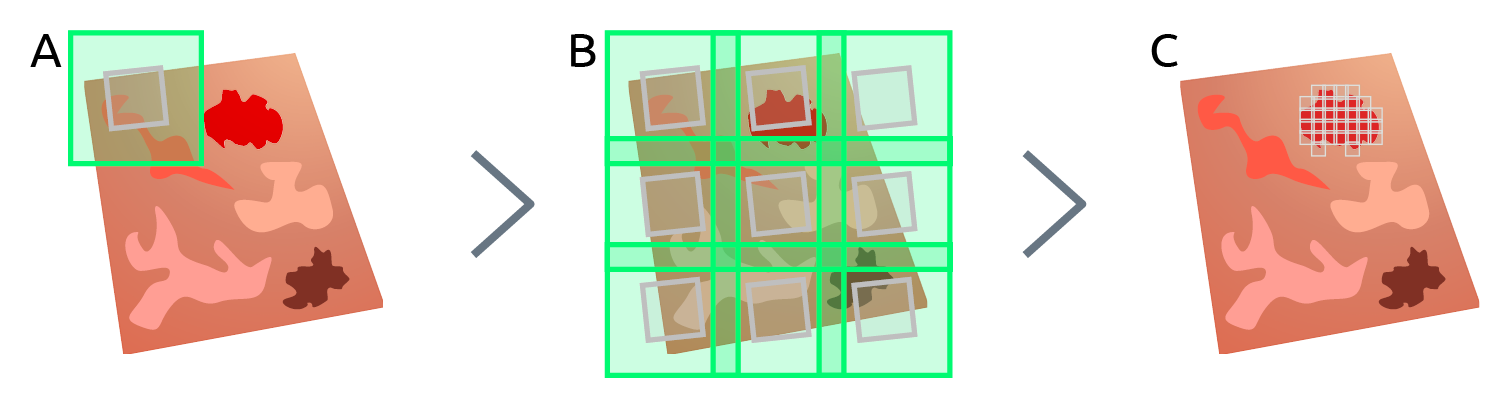
Figure 1 overviews the image acquisition pipeline for integrated array tomography. The process begins by first acquiring a correlative FM and low magnification EM image pair (Figure 1A). A grid of correlative FM, EM image pairs is then acquired across the section with a slight overlap between FM image tiles such that the fluorescence signal of the entire section is fully captured (Figure 1B). To prevent bleaching of the fluorescence due to electron beam irradiation, the FM image tile is always acquired first and the field of view of the low magnification EM image tile is constrained to fit within the non-overlap region of each FM image tile. Immediately following the acquisition of each correlative image pair, an automated registration procedure is executed to maintain sub 10 nm registration accuracy across the entire section [8]. The grid of FM image tiles can then be surveyed to quickly and reliably identify regions of interest based on fluorescence expression for subsequent EM imaging (Figure 1C). Once the biological feature of interest has been imaged with high resolution EM, the pipeline is then repeated on the next serial section and so on until all the sections have been imaged.
Figure 1. Illustration of imaging pipeline for integrated array tomography. First a single (A) then grid (B) of low magnification, correlative FM-EM image pairs is acquired across the section. Regions of interest are then identified based on fluorescence expression for follow-up high magnification EM imaging (C).
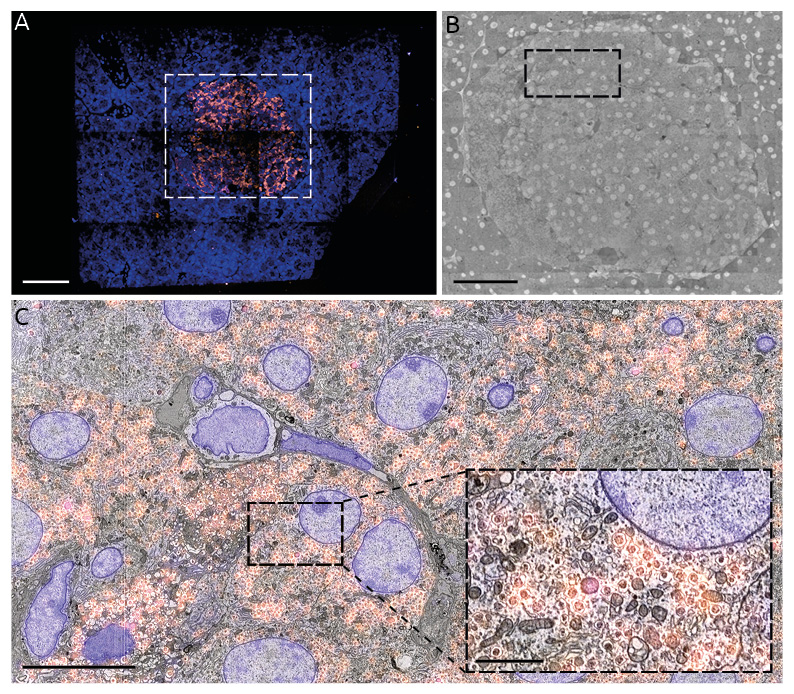
Figure 2 shows the results of the integrated array tomography workflow applied to a single section of rat pancreas tissue labelled for insulin with Alexa 594 (orange) and a Hoechst counterstain (blue). The approximately 1 mm2 tissue section is fully imaged with the fluorescence microscope, revealing the insulin-producing islet of Langerhans in stark contrast (Figure 2A). The approximately 300 um diameter islet (~10% area of the full section) is then fully captured with high magnification (5 nm/px) EM. The high magnification EM images are stitched together [9] and registered to the low magnification EM tiles. The high magnification EM tiles can then be correlated to the FM images via the registration metadata collected from the automated alignment procedure run immediately following the CLEM acquisition.
Figure 2. Full section, correlative FM-EM images of 80 nm rat pancreas tissue following integrated array tomography. (A) FM images. (B) Composite EM image. (C) Correlative overlay. Scale bars: 100 μm (A & B); 10 μm (C); 2μm (C inset).
By limiting the inherently slower, high resolution EM imaging to only those regions revealed by fluorescence expression, we are able to realize considerable gains in throughput. While ROI-based volume reductions are dependent on both the biological specimen and question at hand, the 10% reduction in imaging volume demonstrated here accounts for a directly proportional reduction in time. Additional gains in throughput are realized by virtue of using an integrated microscope. A shared translation stage and software facilitate navigation, while the need for intermediate sample preparation is removed—greatly reducing overhead costs. Further advances in automation, such as machine-learning-assisted ROI detection, have the potential to bring about even greater speed ups, paving the way for high throughput, volume CLEM.
- References
[1] J. Kornfeld and W. Denk, Curr. Opin. Neurobiol. 50 (2018), pp. 261–267.
[2] A. L. Eberle et al., J. Microsc. 259 (2015), pp. 114–120.
[3] Y. Ren and P. Kruit, J. Vac. Sci. Technol. B 34 (2016), p. 06KF02.
[4] W. Yin et al., bioRxiv (2019), p. 791889.
[5] D. G. C. Hildebrand et al., Nature 545 (2017), pp. 345–349.
[6] M. A. Karreman et al., Methods in Cell Biology 140 (2017), pp. 277–301.
[7] J. Delpiano et al., J. Microsc. 271 (2018), pp. 109–119.
[8] M. T. Haring et al. Sci. Rep. 7 (2017).
[9] K. Khairy, G. Denisov, and S. Saalfeld arXiv preprint (2018) arXiv:1804.10019.