Label-free Raman microspectroscopy to study bacteriophage infections
- Abstract number
- 1394
- Event
- Virtual Early Career European Microscopy Congress 2020
- Presentation Form
- Submitted Oral
- DOI
- 10.22443/rms.emc2020.1394
- Corresponding Email
- [email protected]
- Session
- LSA.1 - Label-free life science imaging
- Authors
- Indra Monsees (1), Prof. Dr. Alexander J. Probst (1)
- Affiliations
-
1. Group of Aquatic Microbial Ecology, Universität Duisburg-Essen
- Keywords
Phage, Raman, single-cell, Virus
- Abstract text
We used Raman microspectroscopy to study single unlabeled bacterial cells of Pseudomonas sp. during infection by the bacteriophage phi 6. Our results indicate a significant shift within the Raman spectra of putatively infected cells due to viral infection. The results of this study lay the foundation for correlative approaches for studying viral infection of not-yet cultured microorganisms in environmental samples.
Microscopic identification of microbial cells infected by phage is challenging and only one complex method is currently available [1]. Here, we propose to differentiate phage-infected cells from non-infected cells via Raman microspectroscopy, an emerging technology in natural sciences, which is based on the rare event of inelastic scattering of photons and molecules. Basis for our hypothesis is a study by Huang et al. showing that Raman spectra of unlabeled bacteria allow differentiation of cells between their species level and the respective growth phases [2] as well as the characterization of viral particles via Raman spectroscopes long before Raman microspectroscopes were commercially available [3]. The aim of this work was to find marker Raman shifts to spot viral infections in bacterial pure cultures using multivariate data analysis.
Pseudomonas sp. DSM 21482 was grown in TSB medium and infected with phage phi 6. Before infection and after lysis, samples for Raman spectroscopy were taken, fixed, dehydrated and stored at -20 °C until spectral acquisition using a Renishaw inVia™ confocal Raman Microscope. The acquired spectra were analyzed using the R package Micro Raman [4] using ordination analyses, hierarchical clustering and Monte Carlo-based permutation procedures (multi response permutation procedure, MRPP). Spectral differences were analyzed using contrast plots.
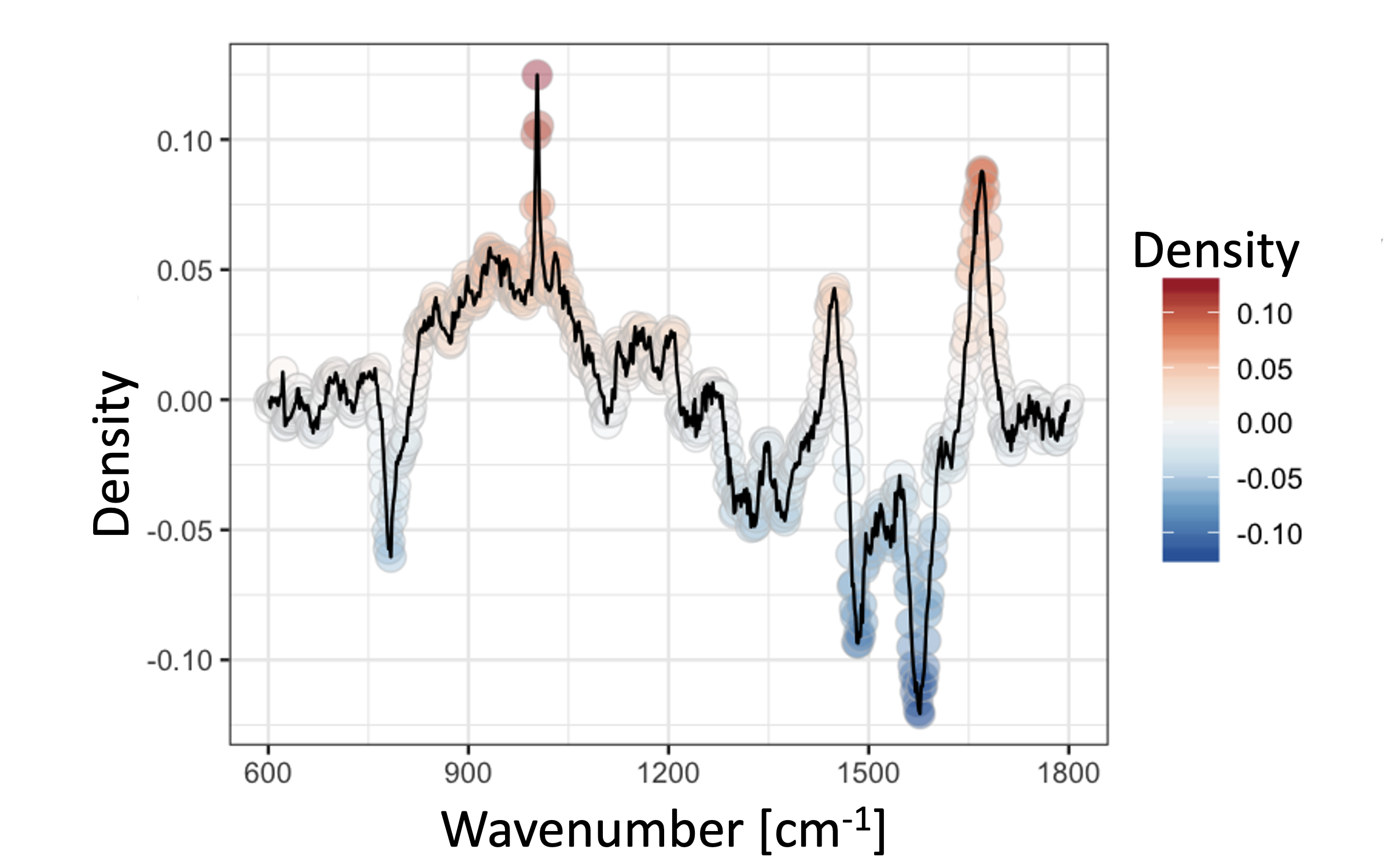
We used two separate Pseudomonas sp. cultures to test our hypothesis, whereas one of them was infected with bacteriophage phi 6. While initially little difference was observed between the two cultures (chance corrected within-group agreement A = 0.0093 p = 0.002), a strong divergence of certain cells after infection was observed (A = 0.06202 p = 0.001). In detail, these cells showed a greater abundance in nucleic acids and lower abundance in proteins. We conclude that these cells are heavily infected and the viral reproduction of its genome causes these shifts in the nucleic acid abundance.
Figure 1 Contrast plot for Raman spectra [4] of the Pseudomonas cells during lysis, comparing the possible infected cells with all other cells of this time point.
Raman spectroscopy is a powerful tool to differentiate uninfected from infected microbial cells. Furthermore, it has a high potential for providing key information for detecting viral infections in environmental samples, based on a change in the ratio of the nucleic acid and protein band intensities, when combined with other microscopy techniques.
- References
[1] Hochstein RA, Amenabar MJ, Munson-McGee JH, et al, Journal of virology 90 (2016) p.3458.
[2] Huang WE, Griffiths RI, Thompson IP, et al, Analytical Chemistry 76 (2004) p.4452.
[3] Li T, Bamford DH, Bamford JKH, et al, Journal of Molecular Biology 230 (1993) p.461.
[4] García-Timermans C, Rubbens P, Kerckhof F-M, et al, Journal of Microbiological Methods 151 (2018) p.69.